🎄12 Days of HPC 2022
Jingle bells, stabbing cells, stealing their mRNA, turn them red, to check they're injected, then come back another day...
During the month of December we’re featuring blog posts from researchers from across the University of Leeds showcasing the fantastic work they do using our High Performance Computing system. Follow us @RC_at_Leeds to keep up to date with our 12 days of HPC blog series.
What’s your name?
Georgette Tanner
What department do you work in?
Leeds Institute of Medical Research at St James’s
What research question are you trying to answer?
Our group is trying to find more effective ways of treating Glioblastoma, the most common and deadly form of brain cancer in adults. Current standard treatment with surgery, chemotherapy and radiation result in the tumours developing resistance and growing back in practically all patients, causing a median survival of just 15 months. We want to understand what it is that enables the tumour cells to develop resistance, in order to develop new treatments that overcome it.
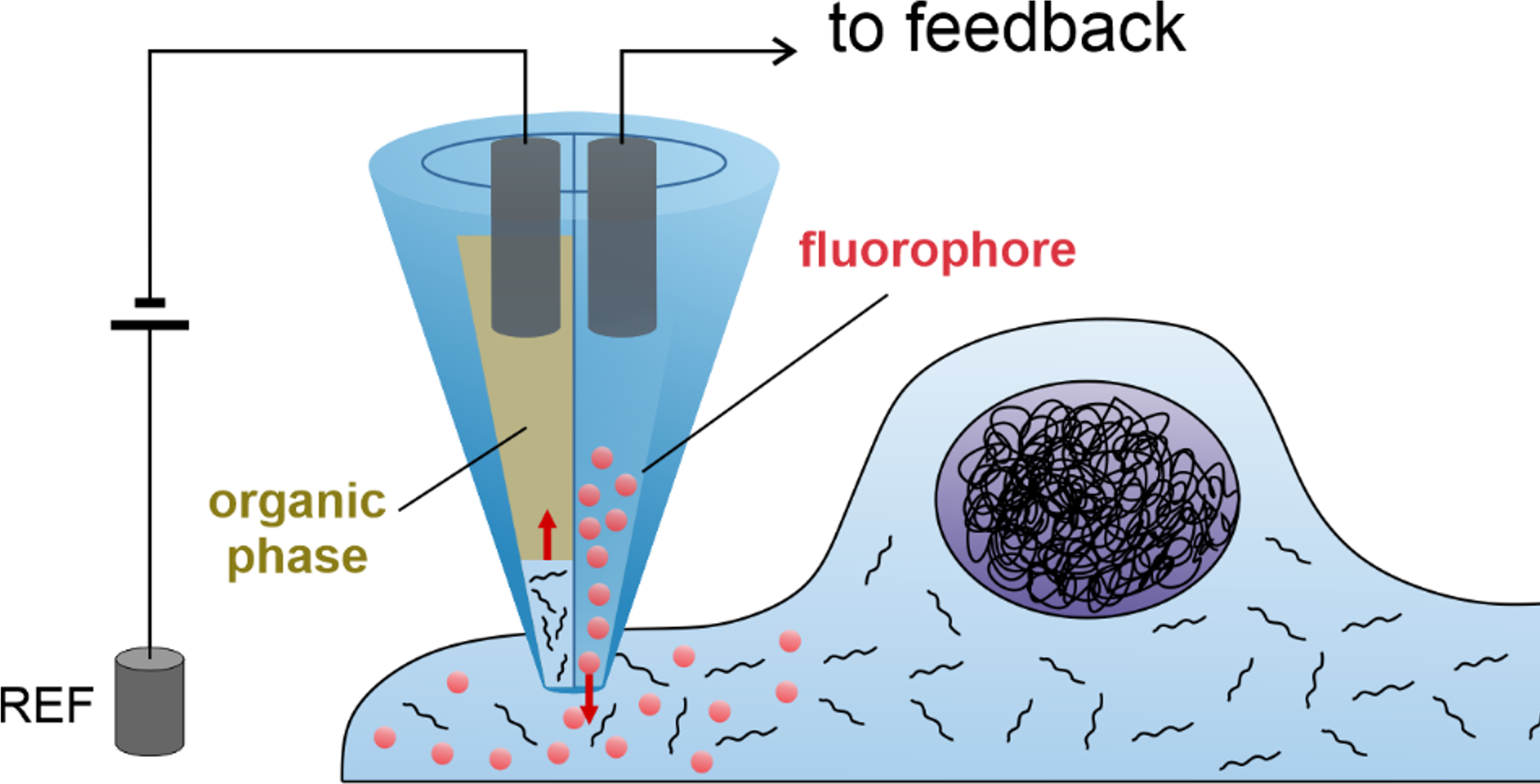
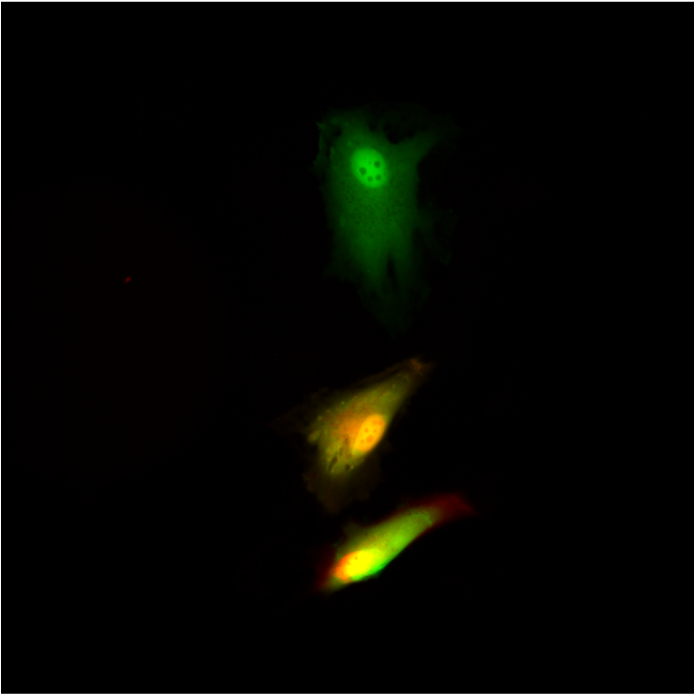
Single cell techniques are commonly used in medical research to characterise the biology of individual cells, but due to the need to kill the cell in the process, they only allow a snapshot of a cell at a single time point. We (Lucy Stead’s Glioma Genomics group) have therefore teamed up with Paolo Actis’ group in the School of Electronic and Electrical Engineering, who have developed a method to take a nano-biopsy of a cell, without killing it. These biopsies sample the cells’ mRNA, which can then be sequenced, allowing us to know what genes are switched on, and also simultaneously injects a red flurophore to confirm that the nano-pipette was inside the cell. Using this technique, Fabio Marcuccio and Chalmers Chau have biopsied glioblastoma cells before treating them with chemotherapy and radiation, and then going back to the same cells and taking another biopsy. This longitudinal approach allows us to identify how individual cells respond to treatment, with insights not previously achievable using standard single cell snapshot approaches.
The final stage in the process, analysing the sequenced mRNA from the biopsies, is where the uni’s HPCs and I are involved. I use this data to try to identify what genes are switched on in each of the biopsied cells, and to then characterise how this changes from the pre- to post-treated cells.
What tools or technologies do you use in your research? (Programming languages, packages, APIs)
The most important methods I use are STAR for aligning the mRNA to a reference genome, followed by the R packages Seurat and DESeq2 for further analyses.
How does HPC help your research?
Billion’s of fragments of mRNA from hundreds of biopsied cells need matching up to a reference human genome in order to know what genes they represent, thereby providing estimates of how strongly genes are switched on. This is followed by various statistical approaches to uncover differences between biopsies. All this requires a lot of computational power which wouldn’t be feasible without the HPC.
What is the potential impact of your research?
Glioblastoma is one of the worst cancer diagnoses a patient can receive. Unlike many cancers, prognoses in glioblastoma have seen no improvements for decades, with just 5% of patients surviving more than 5 years. New treatments are therefore sorely needed. Nano-biopsying glioblastoma cells to see how they respond to treatments will help us to achieve that.
In your personal opinion what’s the coolest thing about your research?
Andy Weir (“The Martian” author) describes a very similar nano biopsy technique in his science fiction novel “Project Hail Mary”, so we’re making science fiction a reality.