🎄12 Days of HPC 2021
Making mRNA translation investigation more accessible on ARC4
During the month of December we’re featuring blog posts from researchers from across the University of Leeds showcasing the fantastic work they do using our High Performance Computing system. Follow us @RC_at_Leeds to keep up to date with our 12 days of HPC blog series.
What’s your name?
Elton Vasconcelos
What department do you work in?
LeedsOmics (FBS & FMH)
What research question are you trying to answer?
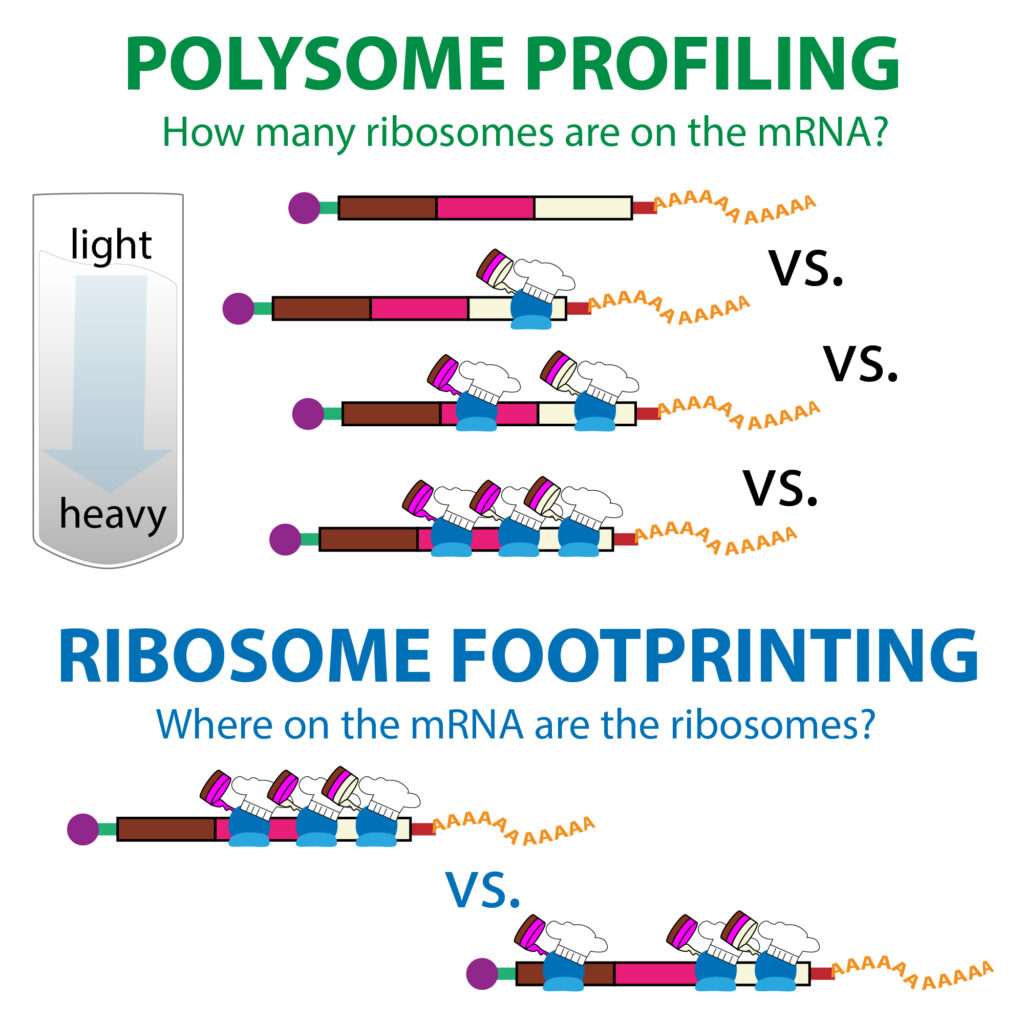
Are there differences in the translational control for different sets of mRNAs?
What tools or technologies do you use in your research? (Programming languages, packages, APIs)
BASH, PERL, R, Python, Java, C++, MySQL, plus dozens of bioinformatics tools, specifically “RiboViz” on this blogspot.
How does HPC help your research?
Providing 24/7 access to our great ARC clusters (3 and 4) which I routinely use to run all my analyses. In addition, there’s a great and talented IT Research team promptly able to help us out through our open tickets.
What is the potential impact of your research?
Better understanding of fundamental molecular biology aspects of the cell, which can be further applied to diseases’ prevention and treatment.
In your personal opinion what’s the coolest thing about your research?
Being able to process and interpret biological big data (omics).
In your opinion, what is the ultimate Christmas song?
Wonderful Christmastime by John Pizzarelli
Postscript
RiboViz is a new and continuously developed tool for Ribosome Profiling data analyses, created by Dr Ed Wallace team @ UoEdinburgh. It has a very complicated and time-consuming installation process. With the help of our IT Research team, I was able to create a singularity container for it on ARC4, which UoLeeds researchers (specially Dr Julie Aspden’s group @ FBS) can benefit of its usage through a fast access and execution through a simple singularity shell -H /nobackup/username /path/to/ribovizEVarc4.sif command on ARC4. Please contact me if you want to run it. :)